Basic Record Linkage with Parallel Processing
When dealing with data from different sources, whether the data are from surveys, internal data, external data vendors, or scraped from the web, we often want to link people or companies across the datasets. Unfortunately, we almost never start with perfectly clean data.
Even when using structured data, people make absent minded mistakes like mixing up letters in names, individual values are recorded incorrectly, and measurement error affects the results. Countless things can happen before we get to dig into the data. Sometimes, perhaps even more frustratingly, different sources just use different names for the same entity (Last.FM vs. LastFM, J.P. Morgan Chase vs. JP Morgan Chase NA, or Robert Griffin 3 vs. Robert Griffin III).
These little discrepancies make it impossible to simply merge the data together on the unique identifiers. Trying to link individual or company data across sources is called record linkage, and it can be extremely time consuming.
Fortunately, we can make this task significantly faster by parallelizing the code. In this post, I’ll walk through basic record linkage (based on string similarity) with and without parallel processing. Then, I’ll measure the speed improvement from parallelizing and discuss why it varies with the size of the data.
Getting Some Data
First things first. I need a toy dataset to play with. I’ll download a list of company names from the SEC, as these are the types of entities that are commonly misspelled in datasets I use.
import requests
import bs4
import multiprocessing as mp
import numpy as np
import time
from difflib import SequenceMatcher
sec_list_html = requests.get('https://www.sec.gov/rules/other/4-460list.htm').content
sec_list_soup = bs4.BeautifulSoup(sec_list_html, 'lxml')
companies_list = sec_list_soup.find_all('tr')[1:-1]
companies_list_clean = [str(x.text.strip()).lower() for x in companies_list]
Let’s take a quick look.
companies_list_clean[:5]
['3com corp',
'3m company',
'a.g. edwards inc.',
'abbott laboratories',
'abercrombie & fitch co.']
Looks good.
I should probably be happy about this, but I somehow couldn’t find a list of company names with spelling errors. So I’ll need to create a synthetic dataset with mistakes by randomly switching a few of the letters in the company names around.
Creating Synthetic Data
I’ll define a function switch_3_characters_randomly to make my synthetic list of company names.
def switch_3_characters_randomly(name, seed = None):
if seed:
np.random.seed(seed)
name_split = list(name)
flip_indices = np.random.choice(len(name), 3, replace = False)
a, b, c = flip_indices[0], flip_indices[1], flip_indices[2]
name_split[a], name_split[b], name_split[c] = name_split[c], name_split[b], name_split[a]
return ''.join(name_split)
Does it work?
synthetic_companies_list = map(switch_3_characters_randomly, companies_list_clean)
print synthetic_companies_list[:5]
print companies_list_clean[:5]
['mco3 corp', '3m copmany', 'a.g. edwards inc.', 'abbottrlabo atories', 'abercro bie & fitchmco.']
['3com corp', '3m company', 'a.g. edwards inc.', 'abbott laboratories', 'abercrombie & fitch co.']
Perfect! Looks like it would be a nightmare to try and merge two datasets with these lists as their company name variables. It’s time for fuzzy matching.
Fuzzy Matching
There are many very smart algorithms for record linkage (William Cohen at Carnegie Mellon has some great information about smart algorithms on his website).
But, since I’m using a toy example, I’ll implement a basic record linkage algorithm that I’d argue is still the most commonly used. This algorithm will calculate the number of matching sequences in two strings and the corresponding similarity ratio. For information about the math behind the similarity ratio, check out the difflib documentation.
First, I’ll define a function to get the similarity ratio of two company names.
def match_ratio(name1, name2):
s = SequenceMatcher(None, name1, name2)
return s.ratio()
Next, I’ll define a function to “fuzzy match” (matching based on the similarity ratio) an individual synthetic company names with the real company names. I’ll decide that a similarity ratio > 0.75 constitutes a possible match. By using a flag variable, we can make sure our output match_list has a value for every company checked (even if no match is found). This may or may not be desirable depending on the goal.
def get_basic_fuzzy_matches(synthetic_name, threshold = 0.75):
match_list = []
flag = None
for name1 in companies_list_clean:
ratio = match_ratio(name1, synthetic_name)
if ratio > threshold:
if not flag:
flag = 1
match_list.append((synthetic_name, name1, ratio))
if not flag:
match_list.append((synthetic_name, None, 0))
return match_list
Let’s do a quick test run to see if the functions are working. I’ll map our function to some of the synthetic company names and see how we did.
matches = map(get_basic_fuzzy_matches, synthetic_companies_list[:50])
matches[:5]
[[('mco3 corp', '3com corp', 0.7777777777777778),
('mco3 corp', 'fmc corp', 0.8235294117647058)],
[('3m copmany', '3m company', 0.9)],
[('a.g. edwards inc.', 'a.g. edwards inc.', 1.0)],
[('abbottrlabo atories', 'abbott laboratories', 0.8947368421052632)],
[('abercro bie & fitchmco.', 'abercrombie & fitch co.', 0.9130434782608695)]]
Not bad! We clearly matched some correctly, but also have some false matches. Fuzzy matching is really useful, but it still requires going through the matched pairs to make sure the potential matches are correct. This takes some time, but way less time than matching by hand.
Parallel Fuzzy Matching
Okay, so we’ve got the function working. How do we parallelize it? By shamelessly using the simple method detailed in this blog post by Chris Kiehl. In Python, we can map functions to data in parallel by using the multiprocessing module. Instead of using Python’s base map function like I did above, I’ll use the map function from multiprocessing.
To actually implement the parallel fuzzy match I will:
- Initialize a multiprocessing.Pool object as
pool - Use the
mapfunction frompooland pass it my fuzzy match function and the synthetic companies list - Close the pool
- Join the output from the pool’s parallel workers
It’s really that simple.
pool = mp.Pool()
pooled_matches = pool.map(get_basic_fuzzy_matches, synthetic_companies_list[:50])
pool.close()
pool.join()
This should be the same as bedore. I’ll make sure they’re the same and look at the first few matches
print pooled_matches == matches
pooled_matches[:5]
True
[[('mco3 corp', '3com corp', 0.7777777777777778),
('mco3 corp', 'fmc corp', 0.8235294117647058)],
[('3m copmany', '3m company', 0.9)],
[('a.g. edwards inc.', 'a.g. edwards inc.', 1.0)],
[('abbottrlabo atories', 'abbott laboratories', 0.8947368421052632)],
[('abercro bie & fitchmco.', 'abercrombie & fitch co.', 0.9130434782608695)]]
Looks like everything works correctly. But why go through this effort? How much faster is it actually?
Timing the Matching
To see how much better the parallel version is, I’ll time how long the fuzzy matching takes for different sized datasets in the standard form and in the parallel form.
indices = map(int, np.linspace(0, len(companies_list_clean), 10))
indices
[0, 105, 210, 315, 420, 526, 631, 736, 841, 947]
Standard Fuzzy Matching
times_list = []
for i in indices[1:]:
t0 = time.time()
matches = map(get_basic_fuzzy_matches, synthetic_companies_list[:i+1])
t1 = time.time()
times_list.append(t1-t0)
Parallel Fuzzy Matching
times_list_pool = []
for i in indices[1:]:
pool = mp.Pool()
t0 = time.time()
pooled_matches = pool.map(get_basic_fuzzy_matches, synthetic_companies_list[:i+1])
pool.close()
pool.join()
t1 = time.time()
times_list_pool.append(t1-t0)
Plotting the Time at Different Sizes
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
f = plt.figure(figsize = (12, 8))
ax = plt.axes()
sns.set(font_scale = 1.25)
plt.plot(indices[1:], times_list, color = 'blue', label = 'Standard Record Linkage')
plt.plot(indices[1:], times_list_pool, color = 'green', label = 'Parallel Record Linkage')
plt.xlabel('Number of Synthetic Companies to Match', size = 15)
plt.ylabel('Time Elapsed (Seconds)', size = 15)
ax.xaxis.set_label_position('bottom')
sns.despine(left = True, bottom = True)
plt.title('Time to Complete Fuzzy Match of Company Names', size = 20)
plt.legend(loc = 'best')
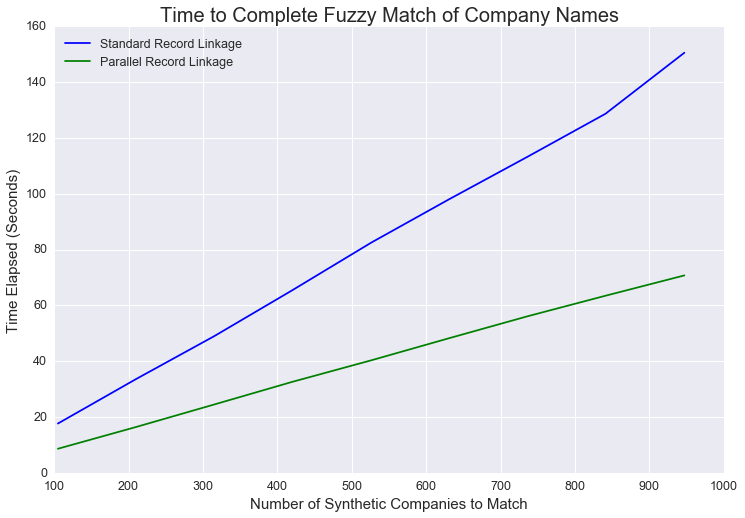
As the size of the dataset increases, the time value of parallel processing increases. With small datasets, parallel processing isn’t much faster. As the dataset gets larger, parallel processing is significantly faster. This makes sense. In order to execute the code in parallel, more actions have to be taken under the hood.
The computer needs to divide the data, store metadata about the divisions, and recombine the data at the end. This is all in addition to mapping the function to the data. This takes non-zero time, so the benefit (in time saved) of parallel programming increases in the size of the dataset.
Conclusion
In this example, we didn’t have that many entities to compare. Our real company dataset has only 947 companies. Parallel record linkage was 2-3 times faster, but we could handle this task without it since it still doesn’t take that long. With more cores to utilize (my laptop only has a couple of cores), the parallel fuzzy match would be even faster and scale more effectively to massive datasets.
This kind of standard record linkage lends itself nicely to being distributed on a computer cluster. In fact, that’s how I would usually run this kind of code.


Leave a Comment